Fast Marching Toolbox Installation tutorial
Contents:
Get the toolbox
Here I provide the basic information to use the codes I provide based on this toolbox. - http://www.mathworks.com/matlabcentral/fileexchange/6110-toolbox-fast-marching
- Note
- DO NOT save the Fast Marching toolbox into the Matlab's toolboxes path. It looks that it does not compile properly in this folder.
More info
System Prerequisites
- Set your Matlab supported compiler.
- For Linux users, install
build-essentialpackage:$ sudo apt-get install build-essential
Compiling the toolbox
- Unzip it in the folder you want and set the Matlab workspace into that folder.
-
Before compiling the mex functions, you will probably need to set up your compiler.
>> mex -setup
And follow the instructions. Working with Ubuntu, the options were:
The options files available for mex are: 1: /usr/local/MATLAB/R2012b/bin/mexopts.sh : Template Options file for building gcc MEX-files 0: Exit with no changesIn my case it wasn't necessary to configure anything. In Windows, this could be problematic... Google will help you :)
-
Run the compile_mex.m script. For that, just type the command:
compile_mex
But you will get the following errors:
Error using mex (line 206) Unable to complete successfully. Error in compile_mex (line 7) mex mex/anisotropic-fm//perform_front_propagation_anisotropic.cppTo solve this error, open the compile_mex.m file and comment lines 7 and 8:% mex mex/anisotropic-fm//perform_front_propagation_anisotropic.cpp % mex mex/anisotropic-fm-feth/fm2dAniso.cppAnd you will find that everything compiles OK now (although you will get many warnings, probably).
Setting the Matlab's Path
The next step is to configure the path of Matlab. For that, you will need to include to your path the following folders:
<path_to_toolbox>/toolbox_fast_marching/data
<path_to_toolbox>/toolbox_fast_marching/
<path_to_toolbox>/toolbox_fast_marching/toolbox
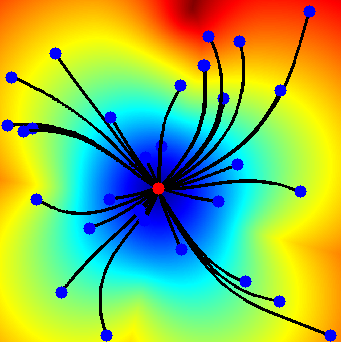
FM toolbox test output
Testing
Go to <path_to_toolbox>/toolbox_fast_marching/tests and run test_fast_marching_2d.m but applying changing line 28 to the following:
end_points = end_points';
Now, it should work an provide an output something like (when clicking more or less in the center of the image):